 Image credit: Chao Hou
Image credit: Chao HouAbstract
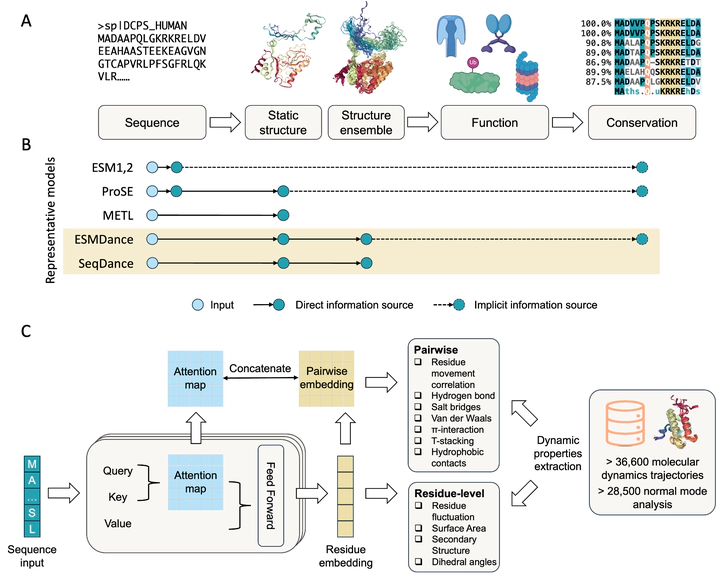
Structural dynamics are fundamental to protein functions and mutation effects. Current protein deep learning models are predominantly trained on sequence and/or static structure data, which often fail to capture the dynamic nature of proteins directly. To address this, we introduce SeqDance and ESMDance, two protein language models trained on dynamic biophysical properties derived from molecular dynamics simulations and normal mode analyses of over 65,100 proteins. SeqDance, trained from scratch, learns both local dynamic interactions and global conformational properties across ordered and disordered proteins. SeqDance predicted dynamic property changes reflect mutation effect on protein folding stability. ESMDance, built upon ESM2 outputs, substantially outperforms ESM2 in zero-shot prediction of mutation effects for designed and viral proteins which lack evolutionary information. Together, SeqDance and ESMDance offer a novel framework for integrating protein dynamics into language models, enabling more generalizable predictions of protein behavior and mutation effect.
Supplementary notes can be added here, including code, math, and images.