Biography
I’m a postdoc at Columbia University in the Shen Lab, where I focus on applying AI for biology. My work centers on developing and interpreting deep learning models, with a particular emphasis on protein language models (pLMs). I’m interested in exploring how protein structural dynamics can be incorporated into deep learning models and how to accurately predict protein fitness landscapes. I developed SeqDance and ESMDance, two pLMs trained on protein dynamics data. I explained the scaling behaviour of pLMs on fitness prediction. I also developed MotifAE for unsupervised discovery of functional motifs from pLMs.
I earned my Ph.D. in Bioinformatics from Peking University in the Li Lab, where I built computational tools for studying phase separation and protein degradation.
I was born in Zhucheng, China, my Chinese name is 侯超. I like basketball, photography, and skiing.
Updated: Nov 2025
- Biological sequence > structure ensemble > function > evolution
- Large language model for biological sequences
- Functional Analysis of Genetic Mutations
- Biomolecular interactions and subcellular localization
- Biomolecular design
Postdoc, 2023.09-
Columbia University
PhD of Bioinformatics, 2020.09-2023.07
Peking University
Bachelor of Medicine and Economics, 2015.09-2020.07
Peking University
Projects
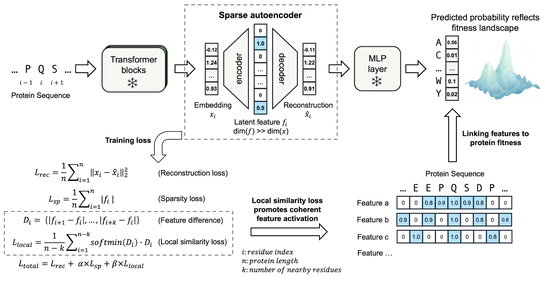
pLM interpretation
Unsupervised discovery of functional motifs from pLM
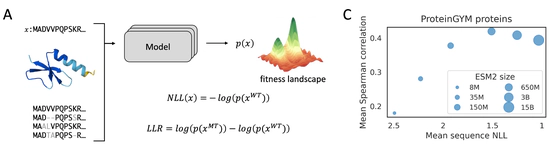
pLM fitness prediction
why larger model do NOT always perform better?
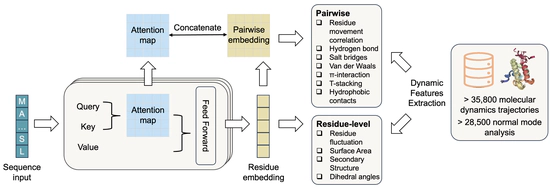
Learn protein dynamics with pLMs
SeqDance/ESMDance trained on Protein Dynamic Properties

Bioinformatics tools for phase seperation
PhaSepDB, PhaSePred and MloDisDB
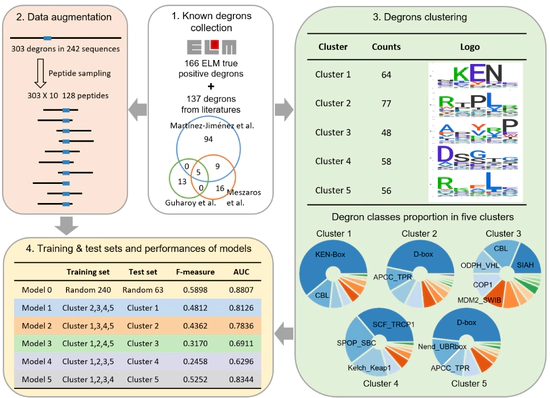
Predicting E3 ligase binding site
Degpred predicts degron and binding E3 via deep learning
Main Publications
Contact
- ch3849@cumc.columbia.edu
- 1130 St Nicholas Ave, New York, NY 10032

