 Image credit: Chao Hou
Image credit: Chao HouAbstract
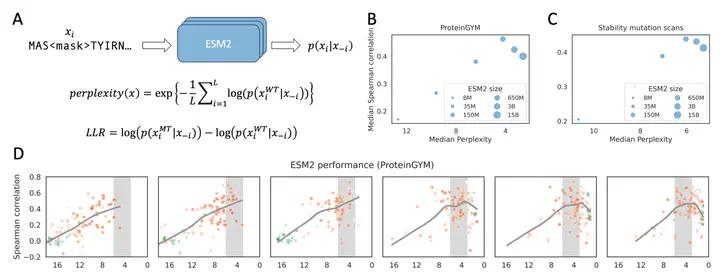
Protein language models (pLMs) can predict mutation effects by computing log-likelihood ratios between mutant and wild-type amino acids, but larger models do not always perform better. We found that the performance of ESM2 peaks when the predicted perplexity for a given protein falls within the range of 3 to 6. Models that yield excessively high or low perplexity tend to predict uniformly near-zero or large negative log-likelihood ratios for all mutations on the protein, limiting their ability to discriminate between deleterious and neutral mutations. Larger models often assign uniformly high probabilities across all positions, reducing specificity for functionally important residues. We also demonstrated how the evolutionary information implicitly captured by pLMs can be linked with the conservation patterns observed in homologous sequences. Our findings highlight the importance of perplexity in mutation effect prediction and suggest a direction for developing pLMs optimized for this application.
Supplementary notes can be added here, including code, math, and images.